Support Vector Machines (Radial Kernel)#
As a classifier, an SVM creates new dimensions from the original data, to be able to seperate the groups along the original features as well as any created dimensions.
The kernel that we choose tells us what constructed dimensions are available to us.
We will start with a linear kernel, which tries to construct hyper-planes to seperate the data.
For 2D, linearly separable data, this is just a line.
We are now going to use a new kernel: RBF, this will create new dimensions that aren’t linear. You do not need to know the details of how this works (that is for later coursework).
We use make_circles because it gives us control over the data and it’s separation; we don’t have to clean or standardize it.
Let’s make some circles#
##imports
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from sklearn.datasets import make_circles
from sklearn.svm import SVC
from sklearn.model_selection import train_test_split
from sklearn.metrics import classification_report, confusion_matrix, roc_curve, roc_auc_score
X,y = make_circles(n_samples = 100, random_state = 3)
## Plot Circles
plt.scatter(X[:,0], X[:,1], c=y)
plt.xlabel(r'$x_0$'); plt.ylabel(r'$x_1$')
Text(0, 0.5, '$x_1$')
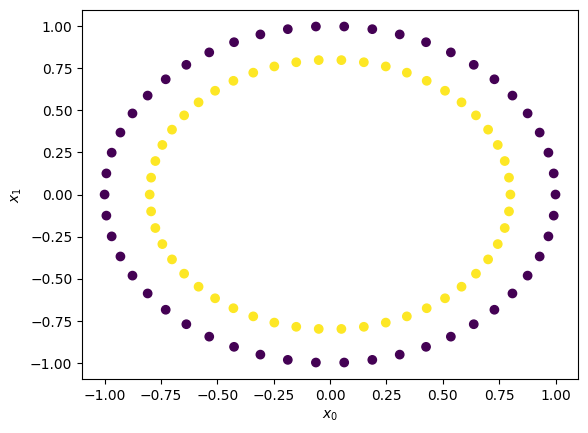
Let’s look at the data in 3D#
fig = plt.figure(figsize = (10, 7))
ax = plt.axes(projection ="3d")
ax.scatter3D(X[:,0], X[:,1], 0, c=y)
<mpl_toolkits.mplot3d.art3d.Path3DCollection at 0xffff4193a9f0>
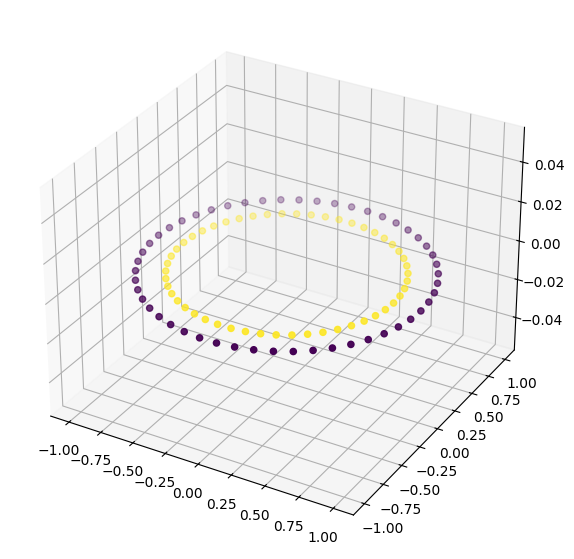
Let’s make a little more data#
X,y = make_circles(n_samples = 1000, random_state = 3)
## Plot Blobs
plt.scatter(X[:,0], X[:,1], c=y)
plt.xlabel(r'$x_0$'); plt.ylabel(r'$x_1$')
Text(0, 0.5, '$x_1$')
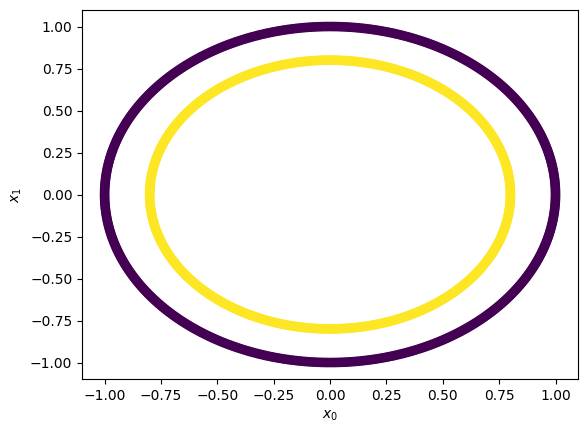
Let’s train up a linear SVM#
This is what we did last class; but now we have split the data
## Split the data
train_vectors, test_vectors, train_labels, test_labels = train_test_split(X, y, test_size=0.25)
## Fit with a linear kernel
cls = SVC(kernel="linear", C=10)
cls.fit(train_vectors,train_labels)
## Print the accuracy
print('Accuracy: ', cls.score(test_vectors, test_labels))
Accuracy: 0.456
Let’s check the report and confusion matrix#
We want more details than simply accuracy
## Use the model to predict
y_pred = cls.predict(test_vectors)
print("Classification Report:\n", classification_report(test_labels, y_pred))
print("Confusion Matrix:\n", confusion_matrix(test_labels, y_pred))
Classification Report:
precision recall f1-score support
0 0.00 0.00 0.00 136
1 0.46 1.00 0.63 114
accuracy 0.46 250
macro avg 0.23 0.50 0.31 250
weighted avg 0.21 0.46 0.29 250
Confusion Matrix:
[[ 0 136]
[ 0 114]]
/home/venv/lib/python3.12/site-packages/sklearn/metrics/_classification.py:1565: UndefinedMetricWarning: Precision is ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, f"{metric.capitalize()} is", len(result))
/home/venv/lib/python3.12/site-packages/sklearn/metrics/_classification.py:1565: UndefinedMetricWarning: Precision is ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, f"{metric.capitalize()} is", len(result))
/home/venv/lib/python3.12/site-packages/sklearn/metrics/_classification.py:1565: UndefinedMetricWarning: Precision is ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, f"{metric.capitalize()} is", len(result))
Let’s look at the ROC curve and compute the AUC#
## Construct the ROC and the AUC
fpr, tpr, thresholds = roc_curve(test_labels, y_pred)
auc = np.round(roc_auc_score(test_labels, y_pred),3)
plt.plot(fpr,tpr)
plt.plot([0,1],[0,1], 'k--')
plt.xlabel('FPR'); plt.ylabel('TPR'); plt.text(0.6,0.2, "AUC:"+str(auc));
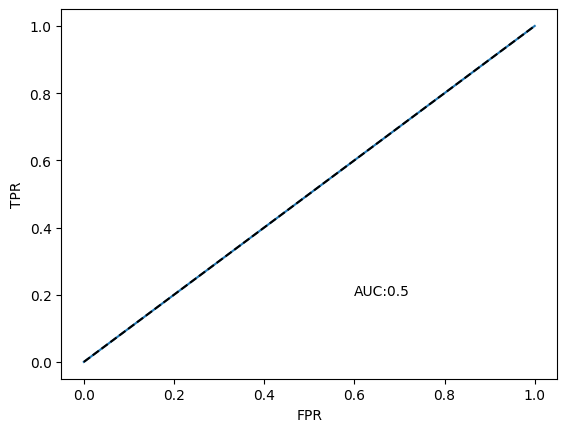
The Linear Kernel Absolutely Failed!#
Let’s use RBF instead and see what happens#
Train the model
Test the model
Evalaute the model: accuracy, scores, confusion matrix, ROC, AUC
Train the model and start evaluating it#
## Fit with a RBF kernel
cls_rbf = SVC(kernel="rbf", C=10)
cls_rbf.fit(train_vectors,train_labels)
## Print the accuracy
print('Accuracy: ', cls_rbf.score(test_vectors, test_labels))
Accuracy: 1.0
Use the model to predict and report out#
## Use the model to predict
y_pred = cls_rbf.predict(test_vectors)
print("Classification Report:\n", classification_report(test_labels, y_pred))
print("Confusion Matrix:\n", confusion_matrix(test_labels, y_pred))
Classification Report:
precision recall f1-score support
0 1.00 1.00 1.00 136
1 1.00 1.00 1.00 114
accuracy 1.00 250
macro avg 1.00 1.00 1.00 250
weighted avg 1.00 1.00 1.00 250
Confusion Matrix:
[[136 0]
[ 0 114]]
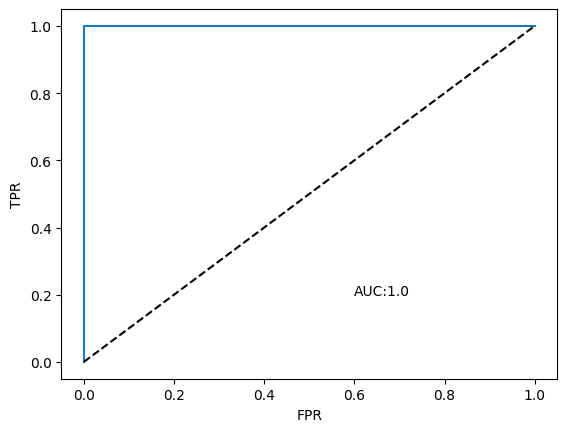
Construct the ROC and the AUC#
## Construct the ROC and the AUC
fpr, tpr, thresholds = roc_curve(test_labels, y_pred)
auc = np.round(roc_auc_score(test_labels, y_pred),3)
plt.plot(fpr,tpr)
plt.plot([0,1],[0,1], 'k--')
plt.xlabel('FPR'); plt.ylabel('TPR'); plt.text(0.6,0.2, "AUC:"+str(auc));
Today#
We are going to use SVM with real data. We are going to use the linear kernel again, but you can change to RBF (it will take much longer to run).
We are also going to introduce hyper-parameter optimization and grid searching (again takes more time)
In the construction of the SVM: cls = svm.SVC(kernel="linear", C=10), C is a hyperparameter that we can adjust. sklearn has a mechanism to do this automatically via a search and find the “best” choice: GridSearchCV.
Please ask lots of questions about what the code is doing today because you are not writing a lot of code today!



