Solution - Exploring data with Pandas#
Today, we will:
Make sure that everyone remembers how to do the basics with
pandas.Do some data analysis with existing data sets.
Make some visualizations of the data.
Notebook instructions#
Work through the notebook making sure to write all necessary code and answer any questions.
Outline:#
Useful imports (make sure to execute this cell!)#
Let’s get a few of our imports out of the way. If you find others you need to add, consider coming back and putting them here.
# necessary imports for this notebook
%matplotlib inline
import pandas as pd
from pandas.plotting import scatter_matrix
1. Review of pandas#
Let’s take a moment to highlight some key concepts. Discuss with your group mates the following prompts and write down a brief definition of each of these concepts.
If you don’t feel like you have good working definitions yet, try doing a quick internet search to see if you can find a definition that makes sense to you.
✅ Question 1: What are the features of a Pandas Series?
Great resource here
✅ Question 2: What are the differences between a DataFrame and a Series?
Nice comparison here
2. Loading and exploring a dataset#
The goal is typically to read some sort of preexisting data into a DataFrame so we can work with it.
Pandas is pretty flexible about reading in data and can read in a variety of formats. However, it sometimes needs a little help. Let’s start with a “toy” dataset, the Iris Data set .
A toy dataset is often one that has a particularly nice set of features or is of a manageable size such that it can be used as a good dataset for learning new data analysis techiques or testing out code. This allows one to focus on the code and data science methods without getting too caught up in wrangling the data. However, data wrangling is an authentic part of doing any sort of meaningful data analysis as data more often messier than not.
Although you will be working with a toy dataset today, you may still have to do a bit of wrangling along the way.
2.1 Getting used to looking for useful information on the internet#
Another authentic part of working as a computational professional is getting comfortable with searching for help on the internet when you get stuck or run into something that is unfamiliar to you. The Python data analysis/computional modeling world can be complicated and is ever-evolving. There is also a large number of publicly available toolsets with varying levels of complexity and nuance. Through practice and experience, we become better computational folks by learning how to search for better, more efficient, and clearer ways to do things.
The Iris data#
The iris data set is pretty straight forward (review the wikipedia link above if you haven’t yet), especially if someone has done all of the data wrangling and cleaning for you. To make it a little more interesting we provide it in a raw form that you might encounter with other data.
✅ Do This: To get started, you’ll need to download the following two files:
https://raw.githubusercontent.com/dannycab/MSU_REU_ML_course/main/notebooks/day-1/iris.data
https://raw.githubusercontent.com/dannycab/MSU_REU_ML_course/main/notebooks/day-1/iris.names
Once you’ve done so, you should have access to the following : iris.data and iris.names. Open them both and discuss what you see. This is a good opportunity to use you favorite text editor or use something new. Feel free to ask your group members or instructors what they prefer to use.
✅ Question 3: Describe the data and the information in the names file. What characteristics are provided there? Perhaps the iris dataset link above will help.
✎ Do This - Erase the contents of this cell and replace it with your answer to the above question! (double-click on this text to edit this cell, and hit shift+enter to save the text). Practice using appropriate Markdown formating to make your answer’s easy to read.
2.2 Reading in a file#
Pandas supports a number of file formats, but one of the most common is the one provided by the read_csv function. Typically, the phrase csv stands for “comma separated values”, a text file format that excel and other spread sheets use. In a typical csv file, each line represents a line in a spreadsheet and each cell value is separated from the next by a comma.
However, you can use other separators as well. Look at the documentation for read_csv.
✅ Do This: Read the iris.data file into your notebook with the appropriate column headers. Display the DataFrame to make sure it looks reasonable.
# replace this cell with code to read in the iris.data file with column headers. Use appropriate headers!
## Note we name the columns in this case because the data file didn't include column names
iris_df = pd.read_csv('https://raw.githubusercontent.com/dannycab/MSU_REU_ML_course/main/notebooks/day-1/iris.data',
names = ['sepal length', 'sepal width', 'petal length', 'petal width', 'species'],
sep = ' ')
iris_df.head()
| sepal length | sepal width | petal length | petal width | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | Iris-setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | Iris-setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | Iris-setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | Iris-setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | Iris-setosa |
2.3 Describing your data#
The .describe() methods tells you about the data. If the data is too big to see (height and width) you can use the two options below to show more of the data. You can change the values to suit your needs.
# expand what is shown (rows and cols) by pandas, any time you change default options -- be thoughtful about your choices!
pd.set_option('display.max_rows', 500)
pd.set_option('display.max_columns', 500)
✅ Do This: Run the .describe() method on the data you read in. If you think about how you might classify the species contained in the dataset based on the four available features, can you discern anything helpful? Put your answer in the cell below.
# Put your code here
iris_df.describe()
| sepal length | sepal width | petal length | petal width | |
|---|---|---|---|---|
| count | 150.000000 | 150.000000 | 150.000000 | 150.000000 |
| mean | 5.843333 | 3.054000 | 3.758667 | 1.198667 |
| std | 0.828066 | 0.433594 | 1.764420 | 0.763161 |
| min | 4.300000 | 2.000000 | 1.000000 | 0.100000 |
| 25% | 5.100000 | 2.800000 | 1.600000 | 0.300000 |
| 50% | 5.800000 | 3.000000 | 4.350000 | 1.300000 |
| 75% | 6.400000 | 3.300000 | 5.100000 | 1.800000 |
| max | 7.900000 | 4.400000 | 6.900000 | 2.500000 |
✎ Do This - Erase the contents of this cell and replace it with your answer. Did you find any useful features? What makes those features useful in classfying the species of the irises?
2.4 Grouping your data#
You can perform operations to group elements using the .groupby() method. The result of the use of .groupby is a GroupBy object. If you were to print it you see only the class instance name as nothing is computed until an operation is performed on the groups, much like other iterators. However, you can use .describe() on the result of a .groupby() call. If you haven’t used this before, you may need to consult the documentation for .groupby()
✅ Do This: Make a GroupBy object using the iris DataFrame and group by the class/species of the data. Then run .describe() on the group result. Now take a look at the data from the group. Thinking again about classification, do you see any data features you could use to classify the data when you look at it this way?
# Put your code here
iris_df.groupby('species').describe()
| sepal length | sepal width | petal length | petal width | |||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | mean | std | min | 25% | 50% | 75% | max | count | mean | std | min | 25% | 50% | 75% | max | count | mean | std | min | 25% | 50% | 75% | max | count | mean | std | min | 25% | 50% | 75% | max | |
| species | ||||||||||||||||||||||||||||||||
| Iris-setosa | 50.0 | 5.006 | 0.352490 | 4.3 | 4.800 | 5.0 | 5.2 | 5.8 | 50.0 | 3.418 | 0.381024 | 2.3 | 3.125 | 3.4 | 3.675 | 4.4 | 50.0 | 1.464 | 0.173511 | 1.0 | 1.4 | 1.50 | 1.575 | 1.9 | 50.0 | 0.244 | 0.107210 | 0.1 | 0.2 | 0.2 | 0.3 | 0.6 |
| Iris-versicolor | 50.0 | 5.936 | 0.516171 | 4.9 | 5.600 | 5.9 | 6.3 | 7.0 | 50.0 | 2.770 | 0.313798 | 2.0 | 2.525 | 2.8 | 3.000 | 3.4 | 50.0 | 4.260 | 0.469911 | 3.0 | 4.0 | 4.35 | 4.600 | 5.1 | 50.0 | 1.326 | 0.197753 | 1.0 | 1.2 | 1.3 | 1.5 | 1.8 |
| Iris-virginica | 50.0 | 6.588 | 0.635880 | 4.9 | 6.225 | 6.5 | 6.9 | 7.9 | 50.0 | 2.974 | 0.322497 | 2.2 | 2.800 | 3.0 | 3.175 | 3.8 | 50.0 | 5.552 | 0.551895 | 4.5 | 5.1 | 5.55 | 5.875 | 6.9 | 50.0 | 2.026 | 0.274650 | 1.4 | 1.8 | 2.0 | 2.3 | 2.5 |
✎ Do this - Erase this and put your answer here. Did you find any useful features? What makes those features useful in classfying the species of the irises? How did using .groupby() help (or not) in finding useful features?
3. Visualizing your data#
We are often limited in understanding our data because it is complex, has many features, or is quite large. In these situations, visualizations (plots, charts, and graphs) can help represent our data in ways that help us gain greater insight into the trends, features, and classes we want to understand.
3.1 Making a scatter matrix#
Rather than just looking at the raw values or exploring basic statistical values for our data, it would be better if we were to visualize the differences. Since the number of features is small, we can use a plotting feature of Pandas called scatter_matrix(). Did you notice that we imported scatter_matrix at the start of this notebook?
✅ Do This: Try calling the scatter_matrix function on your dataframe. Use a marker that you prefer. Look up the documentation for scatter_matrix on the Pandas website for more information! You may need to make the figure bigger to get a good look at it.
Note: There is a similar sort of plot that can be made with the seaborn package, which you may have seen before. If so, do you remember what it is?
# your code here
# Pandas
pd.plotting.scatter_matrix(iris_df,
figsize = (8,8),
alpha = 0.5);
## Seaborn (NOTE THIS SOLVES THE NEXT PROBLEM AUTOMATICALLY)
import seaborn as sns
sns.set_theme(style="ticks")
sns.pairplot(iris_df, hue="species")
<seaborn.axisgrid.PairGrid at 0xffff32a7c8c0>
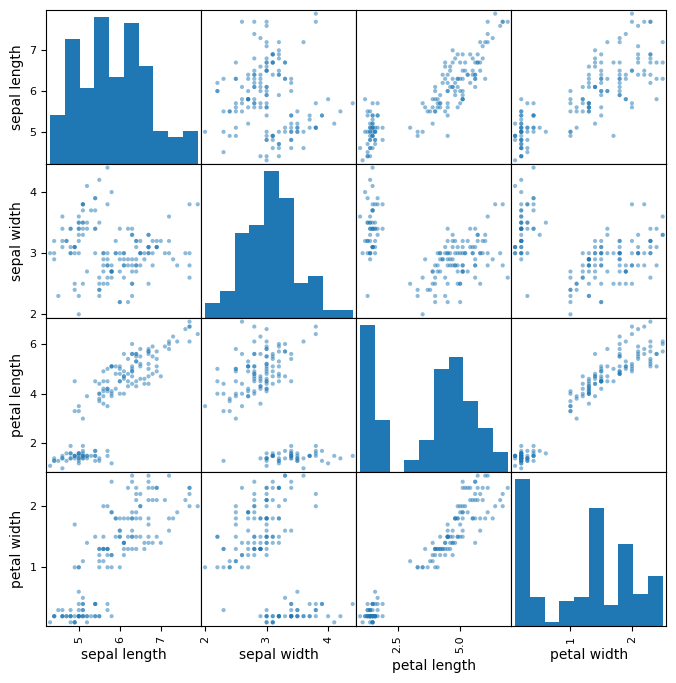
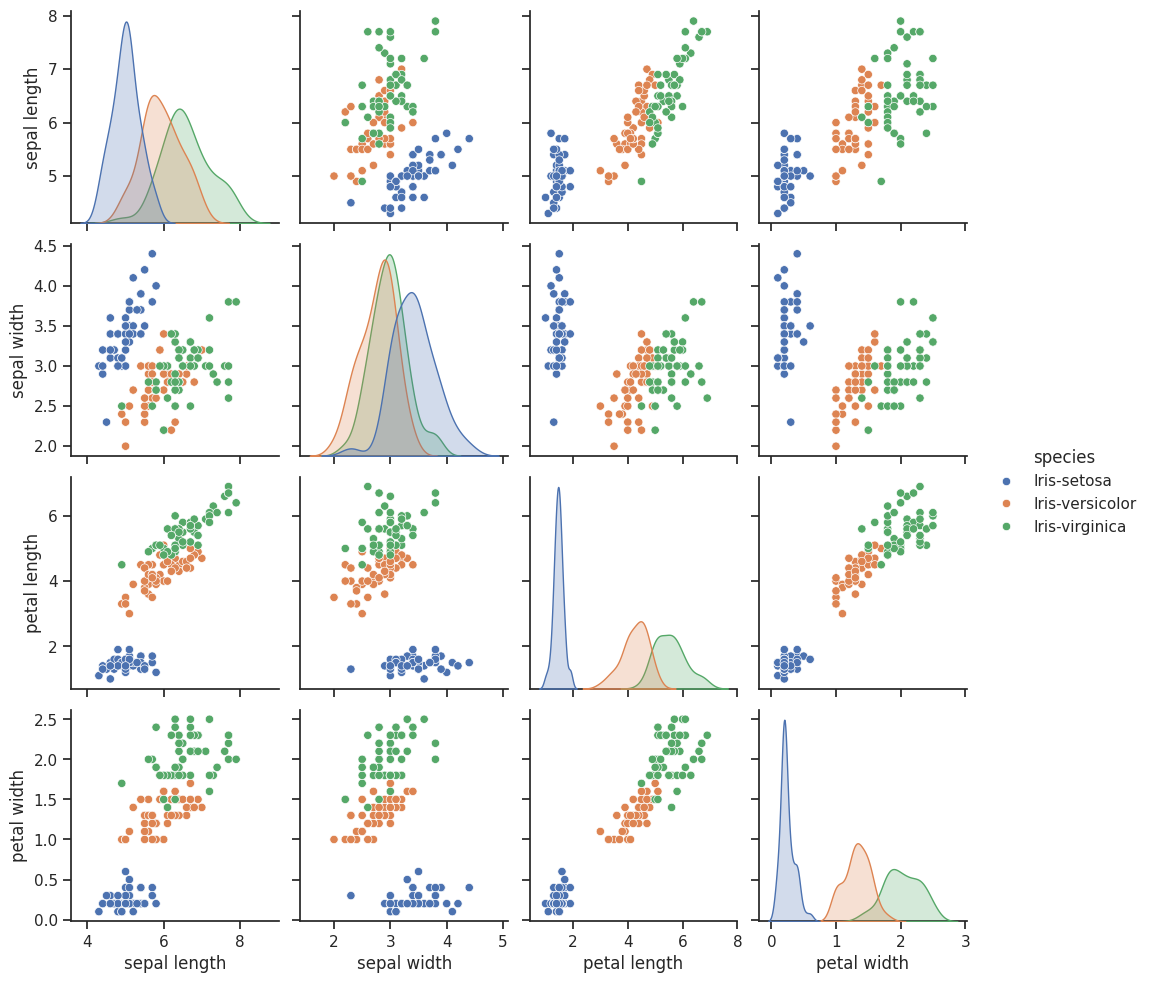
✅ Question 4: Does this visualization help you to determine the features that might be useful for classification? Why or why not?
✎ Do This - Erase the contents of this cell and replace it with your answer to the above question! (double-click on this text to edit this cell, and hit shift+enter to save the text). Practice using appropriate Markdown formating to make your answers easy to read.
3.2 Color coding your data#
The default scatter matrix probably isn’t as helpful as we might hope as it’s impossible to tell which points on the plot represent our classes/sepcies.
We could really use a separate color indication for each dot so we can tell the species apart. Essentially, we need to create an array such that, for each observation in the DataFrame, we can assign a color value to associate it with the appropriate species. It could be just an integer, or it could be one of the standard colors such as “red”.
✅ Do This: Create a new list, array, or Pandas Series object that maps each species to a particular color. Then recreate the scatter matrix using the c argument to give the points in the plot different colors.
# your code here
color = []
color_dict = {'Iris-setosa':'r',
'Iris-versicolor':'g',
'Iris-virginica':'b'}
for x in iris_df['species']:
if x in color_dict:
color.append(color_dict[x])
else:
raise ValueError('No species found in data')
break;
pd.plotting.scatter_matrix(iris_df,
figsize = (8,8),
alpha = 0.5,
c = color);
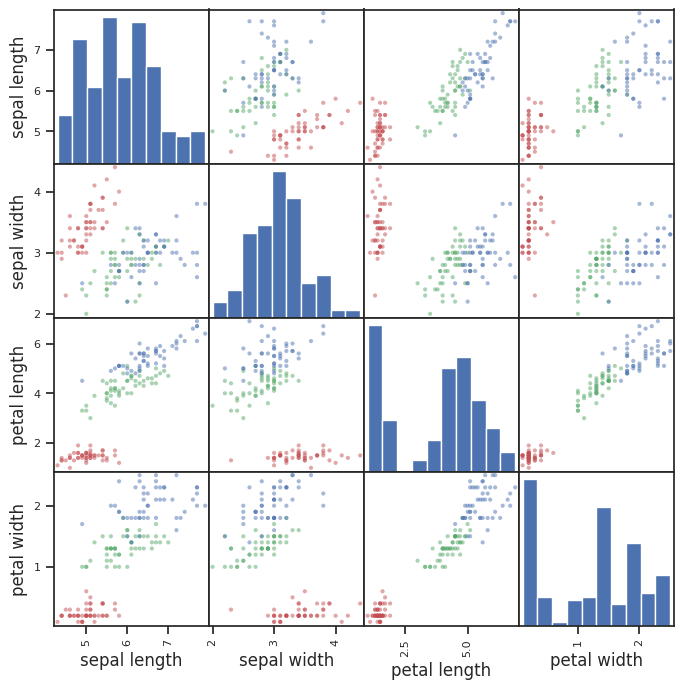
Hope you got something that’s a little more useful! OK, how do we read this now?
The diagonal shows the distribution of the four numeric variables of our example data.
In the other cells of the plot matrix, we have the scatterplots (i.e. correlation plot) of each variable combination of our dataframe.
✅ Question 5: Are you better able to discern features that might be useful in classfying irises? What can you say about the features of each iris species? Can you separate one species easily? If so, using which feature(s)?
✎ Do This - Erase the contents of this cell and replace it with your answer to the above question! (double-click on this text to edit this cell, and hit shift+enter to save the text). Practice using appropriate Markdown formating to make your answers easy to read.
3.3 Separating species of irises#
Now we will use the feature(s) you found above to try to separate iris species. In future parts of the course, we may explore how to do this using models, but for now, you will try to do this by slicing the DataFrame.
✅ Do This: One of these species is obviously easier to separate than the others. Can you use a Boolean mask on the dataframe and isolate that one species using the features and not the species label? Try to do so below and confirm that you were successful.
# Put your code here
## From the graph it looks like we can get Iris-Setosa separated.
iris_df[iris_df['petal length'] < 2] #(only setosa)
#iris_df[iris_df['petal length'] > 2] #(versicolor and virginica)
| sepal length | sepal width | petal length | petal width | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | Iris-setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | Iris-setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | Iris-setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | Iris-setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | Iris-setosa |
| 5 | 5.4 | 3.9 | 1.7 | 0.4 | Iris-setosa |
| 6 | 4.6 | 3.4 | 1.4 | 0.3 | Iris-setosa |
| 7 | 5.0 | 3.4 | 1.5 | 0.2 | Iris-setosa |
| 8 | 4.4 | 2.9 | 1.4 | 0.2 | Iris-setosa |
| 9 | 4.9 | 3.1 | 1.5 | 0.1 | Iris-setosa |
| 10 | 5.4 | 3.7 | 1.5 | 0.2 | Iris-setosa |
| 11 | 4.8 | 3.4 | 1.6 | 0.2 | Iris-setosa |
| 12 | 4.8 | 3.0 | 1.4 | 0.1 | Iris-setosa |
| 13 | 4.3 | 3.0 | 1.1 | 0.1 | Iris-setosa |
| 14 | 5.8 | 4.0 | 1.2 | 0.2 | Iris-setosa |
| 15 | 5.7 | 4.4 | 1.5 | 0.4 | Iris-setosa |
| 16 | 5.4 | 3.9 | 1.3 | 0.4 | Iris-setosa |
| 17 | 5.1 | 3.5 | 1.4 | 0.3 | Iris-setosa |
| 18 | 5.7 | 3.8 | 1.7 | 0.3 | Iris-setosa |
| 19 | 5.1 | 3.8 | 1.5 | 0.3 | Iris-setosa |
| 20 | 5.4 | 3.4 | 1.7 | 0.2 | Iris-setosa |
| 21 | 5.1 | 3.7 | 1.5 | 0.4 | Iris-setosa |
| 22 | 4.6 | 3.6 | 1.0 | 0.2 | Iris-setosa |
| 23 | 5.1 | 3.3 | 1.7 | 0.5 | Iris-setosa |
| 24 | 4.8 | 3.4 | 1.9 | 0.2 | Iris-setosa |
| 25 | 5.0 | 3.0 | 1.6 | 0.2 | Iris-setosa |
| 26 | 5.0 | 3.4 | 1.6 | 0.4 | Iris-setosa |
| 27 | 5.2 | 3.5 | 1.5 | 0.2 | Iris-setosa |
| 28 | 5.2 | 3.4 | 1.4 | 0.2 | Iris-setosa |
| 29 | 4.7 | 3.2 | 1.6 | 0.2 | Iris-setosa |
| 30 | 4.8 | 3.1 | 1.6 | 0.2 | Iris-setosa |
| 31 | 5.4 | 3.4 | 1.5 | 0.4 | Iris-setosa |
| 32 | 5.2 | 4.1 | 1.5 | 0.1 | Iris-setosa |
| 33 | 5.5 | 4.2 | 1.4 | 0.2 | Iris-setosa |
| 34 | 4.9 | 3.1 | 1.5 | 0.1 | Iris-setosa |
| 35 | 5.0 | 3.2 | 1.2 | 0.2 | Iris-setosa |
| 36 | 5.5 | 3.5 | 1.3 | 0.2 | Iris-setosa |
| 37 | 4.9 | 3.1 | 1.5 | 0.1 | Iris-setosa |
| 38 | 4.4 | 3.0 | 1.3 | 0.2 | Iris-setosa |
| 39 | 5.1 | 3.4 | 1.5 | 0.2 | Iris-setosa |
| 40 | 5.0 | 3.5 | 1.3 | 0.3 | Iris-setosa |
| 41 | 4.5 | 2.3 | 1.3 | 0.3 | Iris-setosa |
| 42 | 4.4 | 3.2 | 1.3 | 0.2 | Iris-setosa |
| 43 | 5.0 | 3.5 | 1.6 | 0.6 | Iris-setosa |
| 44 | 5.1 | 3.8 | 1.9 | 0.4 | Iris-setosa |
| 45 | 4.8 | 3.0 | 1.4 | 0.3 | Iris-setosa |
| 46 | 5.1 | 3.8 | 1.6 | 0.2 | Iris-setosa |
| 47 | 4.6 | 3.2 | 1.4 | 0.2 | Iris-setosa |
| 48 | 5.3 | 3.7 | 1.5 | 0.2 | Iris-setosa |
| 49 | 5.0 | 3.3 | 1.4 | 0.2 | Iris-setosa |
✅ Do This: Can you write a sequence of Boolean masks that separate the other two as well? Are these as effective as the first? Can you think of anything you might do to improve the separation?
# your code here
## Looking back maybe petal width can help here
iris_df[(iris_df['petal length'] > 2) & (iris_df['petal width'] < 1.5)] ## We can get pretty close (all versicolor, but one)
#iris_df[(iris_df['petal length'] > 2) & (iris_df['petal width'] > 1.5)] ## Again, we can get pretty close (all virginica, but five)
## Machine Learning Classifiers can combine features and might work better for us
| sepal length | sepal width | petal length | petal width | species | |
|---|---|---|---|---|---|
| 50 | 7.0 | 3.2 | 4.7 | 1.4 | Iris-versicolor |
| 53 | 5.5 | 2.3 | 4.0 | 1.3 | Iris-versicolor |
| 55 | 5.7 | 2.8 | 4.5 | 1.3 | Iris-versicolor |
| 57 | 4.9 | 2.4 | 3.3 | 1.0 | Iris-versicolor |
| 58 | 6.6 | 2.9 | 4.6 | 1.3 | Iris-versicolor |
| 59 | 5.2 | 2.7 | 3.9 | 1.4 | Iris-versicolor |
| 60 | 5.0 | 2.0 | 3.5 | 1.0 | Iris-versicolor |
| 62 | 6.0 | 2.2 | 4.0 | 1.0 | Iris-versicolor |
| 63 | 6.1 | 2.9 | 4.7 | 1.4 | Iris-versicolor |
| 64 | 5.6 | 2.9 | 3.6 | 1.3 | Iris-versicolor |
| 65 | 6.7 | 3.1 | 4.4 | 1.4 | Iris-versicolor |
| 67 | 5.8 | 2.7 | 4.1 | 1.0 | Iris-versicolor |
| 69 | 5.6 | 2.5 | 3.9 | 1.1 | Iris-versicolor |
| 71 | 6.1 | 2.8 | 4.0 | 1.3 | Iris-versicolor |
| 73 | 6.1 | 2.8 | 4.7 | 1.2 | Iris-versicolor |
| 74 | 6.4 | 2.9 | 4.3 | 1.3 | Iris-versicolor |
| 75 | 6.6 | 3.0 | 4.4 | 1.4 | Iris-versicolor |
| 76 | 6.8 | 2.8 | 4.8 | 1.4 | Iris-versicolor |
| 79 | 5.7 | 2.6 | 3.5 | 1.0 | Iris-versicolor |
| 80 | 5.5 | 2.4 | 3.8 | 1.1 | Iris-versicolor |
| 81 | 5.5 | 2.4 | 3.7 | 1.0 | Iris-versicolor |
| 82 | 5.8 | 2.7 | 3.9 | 1.2 | Iris-versicolor |
| 87 | 6.3 | 2.3 | 4.4 | 1.3 | Iris-versicolor |
| 88 | 5.6 | 3.0 | 4.1 | 1.3 | Iris-versicolor |
| 89 | 5.5 | 2.5 | 4.0 | 1.3 | Iris-versicolor |
| 90 | 5.5 | 2.6 | 4.4 | 1.2 | Iris-versicolor |
| 91 | 6.1 | 3.0 | 4.6 | 1.4 | Iris-versicolor |
| 92 | 5.8 | 2.6 | 4.0 | 1.2 | Iris-versicolor |
| 93 | 5.0 | 2.3 | 3.3 | 1.0 | Iris-versicolor |
| 94 | 5.6 | 2.7 | 4.2 | 1.3 | Iris-versicolor |
| 95 | 5.7 | 3.0 | 4.2 | 1.2 | Iris-versicolor |
| 96 | 5.7 | 2.9 | 4.2 | 1.3 | Iris-versicolor |
| 97 | 6.2 | 2.9 | 4.3 | 1.3 | Iris-versicolor |
| 98 | 5.1 | 2.5 | 3.0 | 1.1 | Iris-versicolor |
| 99 | 5.7 | 2.8 | 4.1 | 1.3 | Iris-versicolor |
| 134 | 6.1 | 2.6 | 5.6 | 1.4 | Iris-virginica |
✎ Do This - Erase the contents of this cell and replace it with your answer. How effective your Boolean mask separation of the 3 species? Any ideas for improvements?
4. Physics Data Sets#
We used the famous Iris data set to do this activity. That is because it is well known and clearly documented. Most data sets you will work with will not have the same level of documentation. Let’s try to find some physics data sets that you can read in and plot.
Places to look for data:
Kaggle: Kaggle is a well-known platform for data science and machine learning. It offers a vast collection of datasets contributed by the community. You can search for datasets based on various categories, such as image data, text data, time series data, etc.
UCI Machine Learning Repository: The UCI Machine Learning Repository is a popular resource for machine learning datasets. It provides a diverse collection of datasets from various domains, including classification, regression, clustering, and more.
Google Dataset Search: Google Dataset Search allows you to search for datasets across different domains. It aggregates datasets from various sources, including academic institutions, data repositories, and other websites.
Data.gov: Data.gov is the U.S. government’s open data portal. It provides access to a wide range of datasets covering various topics, such as health, climate, transportation, and more. It’s a valuable resource for finding government-related datasets.
Microsoft Research Open Data: Microsoft Research Open Data is a platform that provides access to diverse datasets collected or curated by Microsoft researchers. It includes datasets from domains like computer vision, natural language processing, and healthcare.
AWS Open Data Registry: AWS Open Data Registry is a collection of publicly available datasets provided by Amazon Web Services (AWS). It hosts a variety of datasets, including satellite imagery, genomics data, and more.
OpenML: OpenML is an online platform that hosts a vast collection of datasets for machine learning research. It also provides tools and resources for collaborative machine learning experimentation.
4.1 Reading data from the internet#
You can pull data directly from the internet using the pandas different read functions. For example read_html can pull data from tables. For example, we can pull the table of all the physical constants from Wikipedia using just a couple lines of Python code.
url = 'https://en.wikipedia.org/wiki/List_of_physical_constants'
tables = pd.read_html(url)
df = tables[0]
df.head()
---------------------------------------------------------------------------
ModuleNotFoundError Traceback (most recent call last)
File /home/venv/lib/python3.12/site-packages/pandas/compat/_optional.py:135, in import_optional_dependency(name, extra, errors, min_version)
134 try:
--> 135 module = importlib.import_module(name)
136 except ImportError:
File /usr/lib/python3.12/importlib/__init__.py:90, in import_module(name, package)
89 level += 1
---> 90 return _bootstrap._gcd_import(name[level:], package, level)
File <frozen importlib._bootstrap>:1387, in _gcd_import(name, package, level)
File <frozen importlib._bootstrap>:1360, in _find_and_load(name, import_)
File <frozen importlib._bootstrap>:1310, in _find_and_load_unlocked(name, import_)
File <frozen importlib._bootstrap>:488, in _call_with_frames_removed(f, *args, **kwds)
File <frozen importlib._bootstrap>:1387, in _gcd_import(name, package, level)
File <frozen importlib._bootstrap>:1360, in _find_and_load(name, import_)
File <frozen importlib._bootstrap>:1324, in _find_and_load_unlocked(name, import_)
ModuleNotFoundError: No module named 'lxml'
During handling of the above exception, another exception occurred:
ImportError Traceback (most recent call last)
Cell In[10], line 2
1 url = 'https://en.wikipedia.org/wiki/List_of_physical_constants'
----> 2 tables = pd.read_html(url)
3 df = tables[0]
5 df.head()
File /home/venv/lib/python3.12/site-packages/pandas/io/html.py:1240, in read_html(io, match, flavor, header, index_col, skiprows, attrs, parse_dates, thousands, encoding, decimal, converters, na_values, keep_default_na, displayed_only, extract_links, dtype_backend, storage_options)
1224 if isinstance(io, str) and not any(
1225 [
1226 is_file_like(io),
(...) 1230 ]
1231 ):
1232 warnings.warn(
1233 "Passing literal html to 'read_html' is deprecated and "
1234 "will be removed in a future version. To read from a "
(...) 1237 stacklevel=find_stack_level(),
1238 )
-> 1240 return _parse(
1241 flavor=flavor,
1242 io=io,
1243 match=match,
1244 header=header,
1245 index_col=index_col,
1246 skiprows=skiprows,
1247 parse_dates=parse_dates,
1248 thousands=thousands,
1249 attrs=attrs,
1250 encoding=encoding,
1251 decimal=decimal,
1252 converters=converters,
1253 na_values=na_values,
1254 keep_default_na=keep_default_na,
1255 displayed_only=displayed_only,
1256 extract_links=extract_links,
1257 dtype_backend=dtype_backend,
1258 storage_options=storage_options,
1259 )
File /home/venv/lib/python3.12/site-packages/pandas/io/html.py:971, in _parse(flavor, io, match, attrs, encoding, displayed_only, extract_links, storage_options, **kwargs)
969 retained = None
970 for flav in flavor:
--> 971 parser = _parser_dispatch(flav)
972 p = parser(
973 io,
974 compiled_match,
(...) 979 storage_options,
980 )
982 try:
File /home/venv/lib/python3.12/site-packages/pandas/io/html.py:918, in _parser_dispatch(flavor)
916 import_optional_dependency("bs4")
917 else:
--> 918 import_optional_dependency("lxml.etree")
919 return _valid_parsers[flavor]
File /home/venv/lib/python3.12/site-packages/pandas/compat/_optional.py:138, in import_optional_dependency(name, extra, errors, min_version)
136 except ImportError:
137 if errors == "raise":
--> 138 raise ImportError(msg)
139 return None
141 # Handle submodules: if we have submodule, grab parent module from sys.modules
ImportError: Missing optional dependency 'lxml'. Use pip or conda to install lxml.
✎ Do This - Find a physics data set of interest and read it in with pandas (from the web). Make plots of the data contained in the file.
## your code here
🛑 You made it to the end!#
Is there anything that we covered today that you’re still not feeling certain about? Talk to your group or check in with an instructor.



